CAT #: 94120091
LeukoStrat ® FLT3 Mutation Assay 2.0 – ABI Fluorescence Detection
Intended Use
The LeukoStrat®FLT3 Mutation Assay 2.0 is an in vitro diagnostic product intended for PCR-based detection of FLT3 activating mutations in patients with acute myelogenous leukemia (AML).
Specifically, the FLT3 Mutation Assay 2.0 can be used to:
- Identify internal tandem duplications (ITD) in the FLT3 gene.
- Identify tyrosine kinase domain (TKD) mutations in the FLT3 gene.
Product Details
-
Summary and Explanation of the Test
Acute myeloid leukemia (AML) in general has a poor prognosis. Assessment of the mutation status of the FLT3 (fms like tyrosine kinase 3) receptor gene in karyotype normal AML is the most important prognostic indicator of disease outcome, which is often substantial, as many studies in AML have shown that the presence of FLT3 activating mutations portends a poor prognosis1, 2. For this reason FLT3 mutation testing is required to stratify disease and determine appropriate treatment options. This LeukoStrat® assay targets regions of the FLT3 gene to identify internal tandem duplication (ITD) mutations and tyrosine kinase domain (TKD) mutations, such as the D835 and I836 mutations. With this assay, DNA is amplified via PCR with fluorophore labeled primers, TKD amplicon is enzymatically digested, and the amplicons are detected via capillary electrophoresis.
This product includes fluorophore-labeled PCR master mixes, along with positive and negative controls for mutant detection via capillary electrophoresis. This assay reliably detects FLT3 mutations comprising greater than or equal to 5% of the total cell population.
-
Principles of the Procedure
Internal Tandem Duplication (ITD) Mutations of FLT3
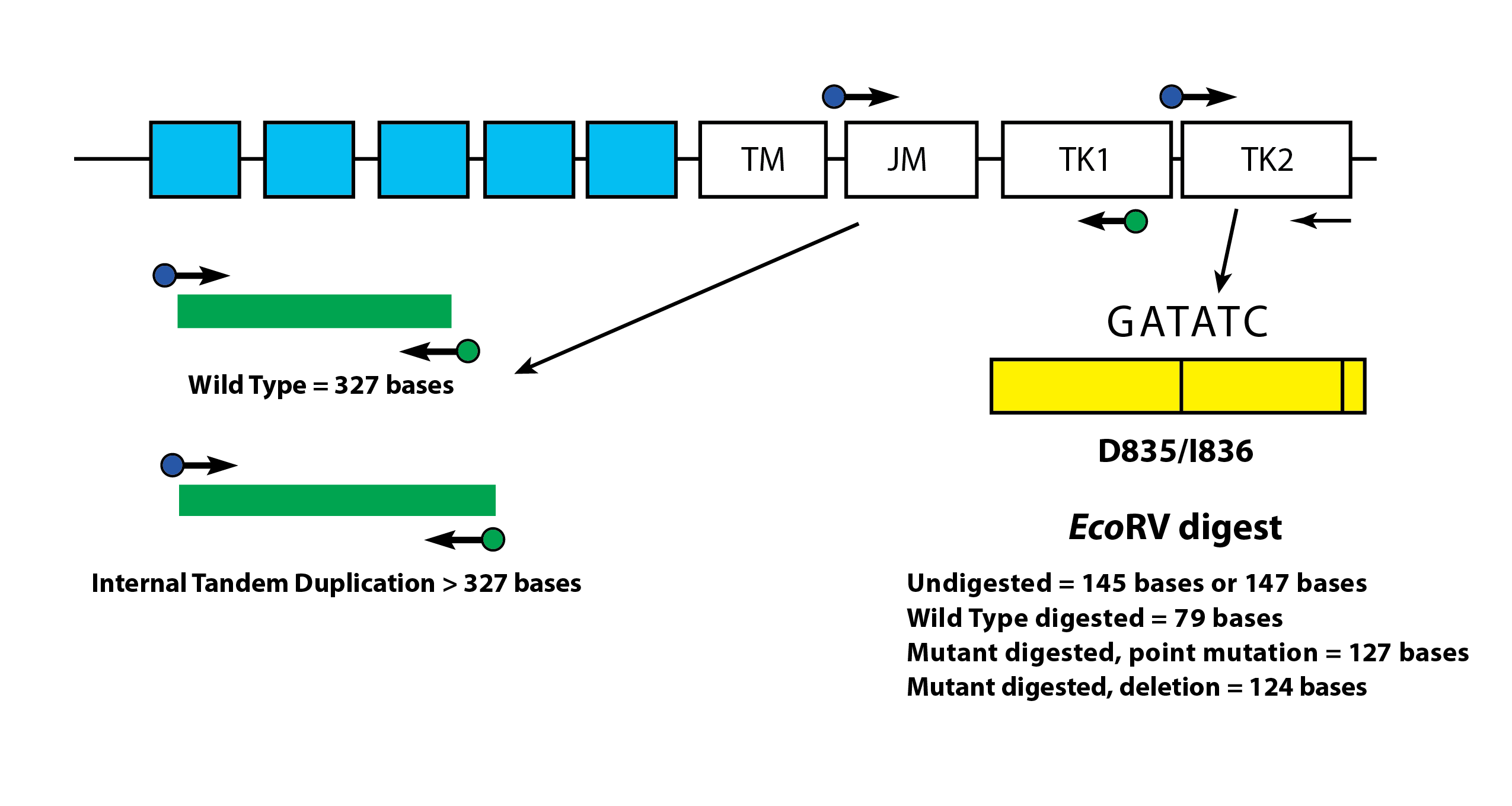
FLT3 ITD or length mutations are caused by duplication and insertion of a portion of the FLT3 gene that includes the region in and around the juxtamembrane (JM) region of the FLT3 gene. These mutations vary in both the location and the length of the inserted duplicated DNA sequence. ITD mutations result in constitutive autophosphorylation and activation of FLT3. Using this assay, FLT3 mutant alleles are differentiated from wild type based on amplicon size as detected via capillary electrophoresis.
Depicted is a representation of the FLT3 JM region and the activating loop of the kinase domain. Green and blue dots with black arrows represent the relative positions of primers that target in and around the JM region for ITD and yellow dots with black arrows represent the relative positions of the primers that target TKD mutations in the activating loop of the kinase domain. The yellow box has vertical black lines that represent the position of the wild-type EcoRV restriction digest sites.
Tyrosine Kinase Domain (TKD) Mutations of FLT3
FLT3 TKD mutations are caused by nucleic acid substitutions that result in a change in the amino acid sequence in this highly conserved catalytic center. TKD mutations, such as D835 and I836, result in constitutive autophosphorylation and activation of FLT32. Wild-type alleles of the FLT3 gene include an EcoRV restriction digest site. When a nucleic acid substitution occurs, the restriction digest recognition site disappears, and the EcoRV endonuclease is unable to identify and digest the DNA at this site. Our tests use primers that lie on either side of the TKD region. The FLT3 target region is amplified using PCR and then an EcoRV restriction digest is performed. One of the PCR primers contains an EcoRV restriction site, so both wild type and mutant alleles are digested. Using this assay, FLT3 mutant alleles are differentiated from wild type based on amplicon size as detected via capillary electrophoresis. Amplicon size further controls for EcoRV digestion.
Differential Fluorescence Detection
Differential fluorescence detection is commonly used to resolve the different-sized amplicon products using a capillary electrophoresis instrument. Primers can be conjugated with several different fluorescent dyes (fluorophors) so that they can produce different emission spectra upon excitation by a laser in the capillary electrophoresis instrument. In this manner, different fluorescent dyes can correspond to different targeted regions. This detection system results in unsurpassed sensitivity, single nucleotide resolution, differential product detection, and relative quantification. In addition, the use of agarose and polyacrylamide gels, as well as the use of carcinogens such as ethidium bromide, can virtually be eliminated. Further, differential detection allows accurate, reproducible and objective interpretation of primer-specific products and automatic archiving of data. Inter-assay and intra-assay reproducibility in size determination using capillary electrophoresis is approximately 1 to 2 nucleotides. This reproducibility and sensitivity coupled with the automatic archiving of specimen data allows for the monitoring, tracking, and comparison of data from individual patients over time.
-
Specimen Requirements
This assay tests genomic DNA from the following sources:
-
Peripheral blood or bone marrow aspirate anti-coagulated with heparin, EDTA, or ACD (stored at 2°C to 8°C for <=7 days).
-
Isolated mononuclear cells that are fresh in an appropriate media such as RPMI (stored for <=7 days), or frozen in an appropriate cryopreservation media (stored indefinitely).
- 500 ng of genomic DNA (stored at 2°C to 8°C or below -15°C and shipped at ambient temperature, cool conditions, or on dry ice).
-
References
1. Murphy, KM. et al., (2003). Detection of FLT3 Internal Tandem Duplication and D835 Mutations by a Multiplex Polymerase Chain Reaction and Capillary Electrophoresis Assay. The Journal of Molecular Diagnostics 5, 96 – 102.
2. Yamamoto, Y. et al., (2001). Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 97, 2434-2439.
Legal Notice
This product is an in vitro diagnostic product; not available for sale or use within North America.
Many of these products require nucleic acid amplification methods such as Polymerase Chain Reaction (PCR). No license under these patents to use amplification processes or enzymes is conveyed expressly or by implication to the purchaser by the purchase of this product.
©2023 Invivoscribe, Inc. All rights reserved. The trademarks mentioned herein are the property of Invivoscribe, Inc. and/or its affiliates, or (as to the trademarks of others used herein) their respective owners.

 Outside North America
Outside North America