CAT #: K4120431
LeukoStrat® CDx FLT3 Mutation Assay
Intended Use
The LeukoStrat CDx FLT3 Mutation Assay is a PCR-based in vitro diagnostic test designed to detect internal tandem duplication (ITD) and tyrosine kinase domain (TKD) mutations D835 and I836 in the FLT3 gene in genomic DNA extracted from mononuclear cells obtained from peripheral blood or bone marrow aspirates of patients diagnosed with acute myelogenous leukemia (AML). The LeukoStrat CDx FLT3 Mutation Assay may be used as a companion diagnostic for the following therapeutic:
In regions where XOSPATA® (gilteritinib fumarate) is available, the LeukoStrat CDx FLT3 Mutation Assay is used as an aid in the assessment of patients with AML for whom XOSPATA (gilteritinib fumarate) treatment is being considered.
In regions where VANFLYTA® (quizartinib hydrochloride) is available, the LeukoStrat CDx FLT3 Mutation Assay is used as an aid in the assessment of patients with FLT3-ITD+ AML for whom VANFLYTA® (quizartinib hydrochloride) treatment is being considered.
The qualitative, non-automated test is for use on the 3500xL or 3500xL Dx Genetic Analyzers.
Product Details
-
Summary and Explanation of the Test
Acute myelogenous leukemia (AML) in general has a poor prognosis. Assessment of the mutation status of the FLT3 (fms related tyrosine kinase 3) receptor gene in karyotype normal AML is the most important prognostic indicator of disease outcome, which is often substantial, as many studies in AML have shown that the presence of FLT3 activating mutations portends a poor prognosis.1,2 The LeukoStrat® CDx FLT3 Mutation Assay targets regions of the FLT3 gene to identify internal tandem duplication (ITD) mutations and tyrosine kinase domain (TKD) mutations, such as the D835 and I836 mutations.
The LeukoStrat® CDx FLT3 Mutation Assay includes reagents, equipment, software and procedures for isolating mononuclear cells and extracting DNA from patient specimens to determine if FLT3 mutations are present. DNA is amplified via PCR and the amplicons are detected via capillary electrophoresis. FLT3 mutation status is determined by the LeukoStrat® CDx FLT3 Software.
-
Principles of the Procedure
Internal Tandem Duplication (ITD) Mutations of FLT3
FLT3 ITD or length mutations are caused by duplication and insertion of a portion of the FLT3 gene that includes the region in and around the juxtamembrane (JM) region of the FLT3 gene. These mutations vary in both the location and the length of the inserted duplicated DNA sequence. ITD mutations result in constitutive autophosphorylation and activation of FLT3.
The LeukoStrat® CDx FLT3 Mutation Assay uses primers that are in and around the JM region. The forward and reverse PCR primers are fluorescently labeled with different fluorophores that serve to confirm the presence of sample signal. Wild-type FLT3 alleles will amplify and produce a product measured at 327±1 bp as measured by this assay, while alleles that contain ITD mutations will produce a product that exceeds 327±1 bp (Figure 1).
Tyrosine Kinase Domain (TKD) Mutations of FLT3
FLT3 TKD mutations are caused by nucleic acid substitutions and/or deletions that result in a change in the amino acid sequence in this highly-conserved catalytic center. TKD mutations, such as D835 and I836 substitutions and deletions, result in constitutive autophosphorylation and activation of FLT3.
Wild-type alleles of the FLT3 gene include an EcoRV restriction digest site. When a nucleic acid substitution occurs, the restriction digest recognition site disappears, and the EcoRV endonuclease is unable to identify and digest the DNA at this site. The LeukoStrat® CDx FLT3 Mutation Assay uses primers that lie on either side of the TKD region. The FLT3 target region is amplified using PCR and then an EcoRV restriction digest is performed. One of the PCR primers is labeled with a fluorophore and the other contains an engineered EcoRV restriction site, so both wild type and mutant alleles are digested. The digestion pattern identifies loss of the normal gene sequence and ensures that digestion occurred. Wild-type alleles of the FLT3 gene yield digestion products of 79±1 bp whereas mutant alleles yield products of 125±1 bp or 127±1 bp from the original undigested amplicon product of 145±1 bp or 147±1 bp as measured by this assay (Figure 1).
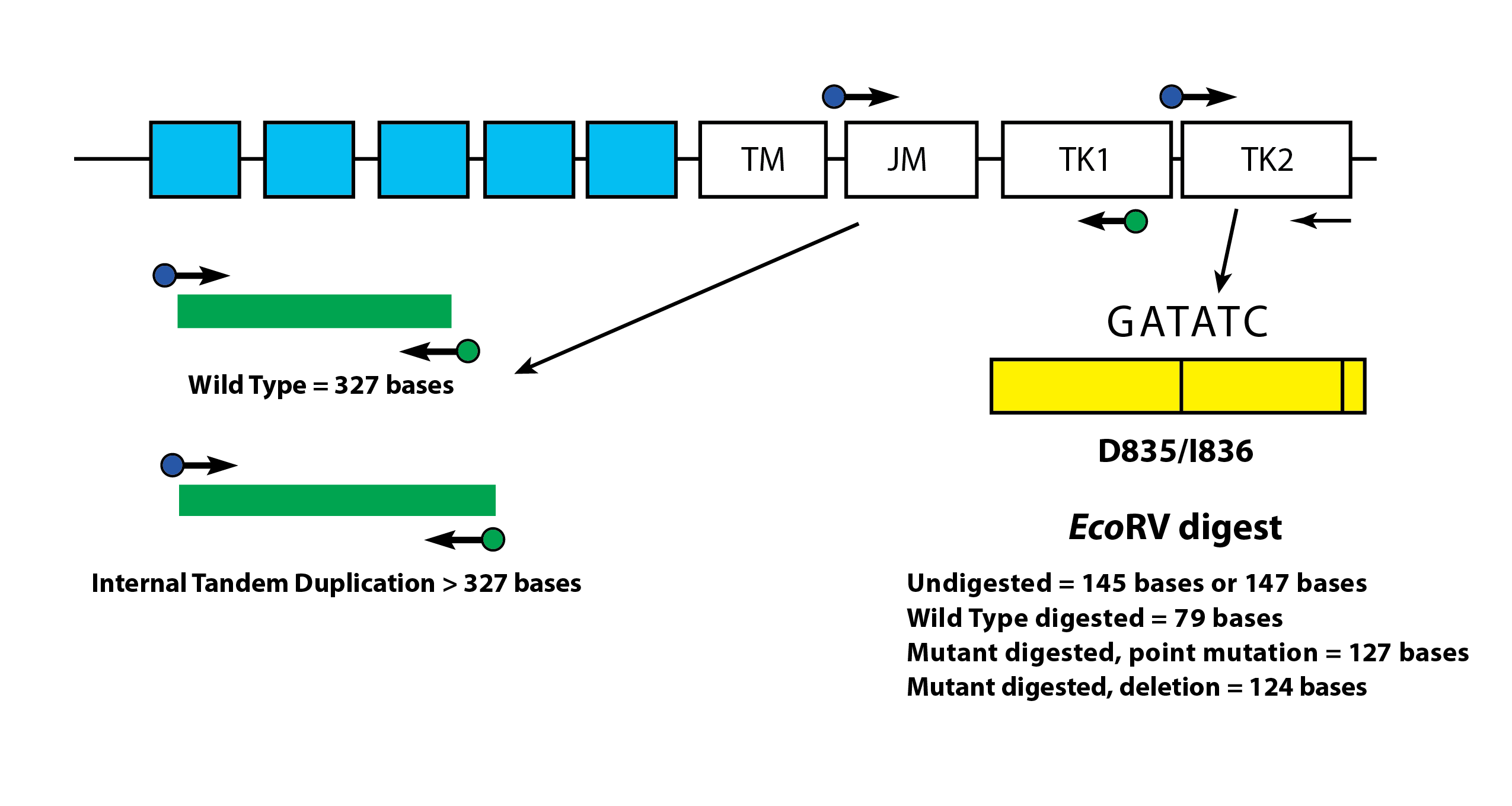
Figure 1: Depicted is a representation of the FLT3 juxtamembrane (JM) region (TM = transmembrane) and the activating loop of the tyrosine kinase (TK) domain. Black arrows represent the relative positions of primers that target in and around the JM region for ITD or the activating loop of the kinase domain for TKD. Colored dots represent fluorophores on labeled primers. The yellow box has vertical black lines that represent the position of the EcoRV restriction digest sites.
Disclaimer
CE-2797 IVD products are intended for in vitro diagnostic use. Not available in North America.
Legal Notice
For Legal Notices related to this product, visit: https://invivoscribe.com/legal-notices/

 Outside North America
Outside North America 
