CAT #: 51010031
IGH Somatic Hypermutation Assay v2.0 for ABI Fluorescence Detection
Assay Use
The Research Use Only IGH Somatic Hypermutation Assay v2.0 is used to identify clonal rearrangements of the immunoglobulin heavy (IGH) chain gene and determine the somatic mutation status of the variable (V) gene sequence and is useful for the study of:
- Identifying clonal rearrangements of the IGH chain gene
- Assessing the extent of somatic hypermutation in the variable region of the immunoglobulin heavy chain gene
- Evaluating new research and methods in malignancy studies
Product Details
-
Summary and Explanation of the Test
BACKGROUND
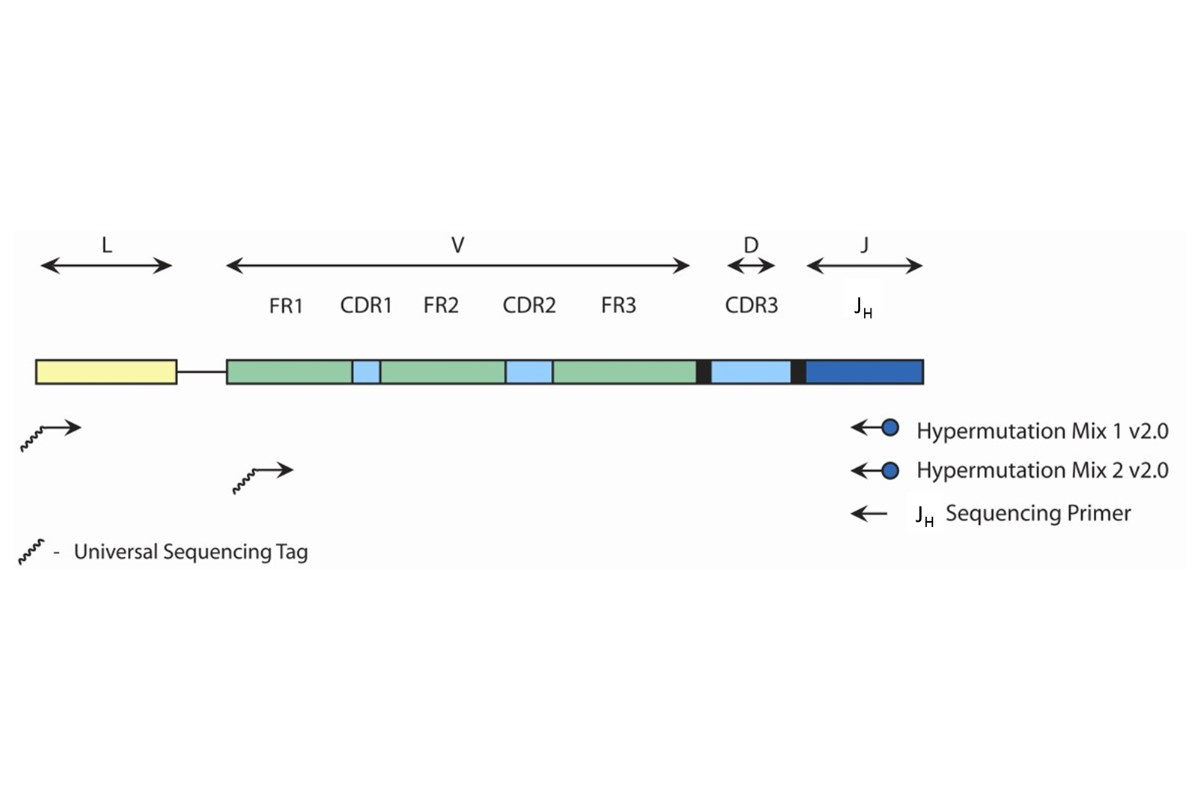
Rearrangements of the antigen receptor genes occur during ontogeny in B and T lymphocytes. These gene rearrangements are unique in length and sequence for each cell. Therefore, polymerase chain reaction (PCR) assays can be used to identify lymphocyte populations derived from a single cell by detecting the unique V-J gene rearrangements present within these antigen receptor loci.1 This PCR-based assay employs multiple consensus DNA primers that target conserved genetic regions within the immunoglobulin heavy chain (IGH) gene. This test is used to detect and sequence the majority of clonal IGH rearrangements from either genomic DNA (gDNA) or complementary DNA (cDNA). Clonal products can be detected using a variety of methods, including gel and capillary electrophoresis.The primers that target the leader (VHL) and framework 1 (FR1) regions have been designed to include a universal sequencing tag at the 5’end. This design allows for bi-directional sequencing of clonal PCR products with just one sequencing-tag specific forward primer and one JH reverse primer. The presence of IGH somatic hypermutation (SHM) is defined as greater or equal to 2% difference from the germline variable (V) gene sequence, whereas less than 2% difference is considered evidence of no somatic hypermutation.
SUMMARY
This test amplifies either gDNA or cDNA that lies between the upstream leader (VHL) or framework 1 (FR1) regions and the downstream joining (J) region of the IGH gene. The test utilizes two (2) different master mixes: Hypermutation Mix 1 v2.0 and Hypermutation Mix 2 v2.0. The Hypermutation Mix 1 v2.0 targets sequences between the leader and joining regions. Therefore the amplicon product(s) span the entire variable (V) region, which contains the FR1, CDR1 (complementarity-determining region 1), FR2, CDR2, FR3 and CDR3 regions. The Hypermutation Mix 2 v2.0 targets sequences between the framework 1 (FR1) and joining (J) regions. The resulting amplicons include a portion of the FR1 region to the downstream J region. Accordingly products do not include the complete FR1 sequence. -
Specimen Requirements
This assay tests extracted and purified gDNA or cDNA derived from RNA. Common sources of gDNA and RNA include:
- 5 cc of peripheral blood, bone marrow biopsy or bone marrow aspirate anti-coagulated with heparin or EDTA (stored at 2ºC to 8ºC and shipped at ambient temperature)
- Formalin-fixed paraffin embedded tissue or slides (stored and shipped at ambient temperature)
Legal Notice
Warranty and Liability
Invivoscribe, Inc. (Invivoscribe®) is committed to providing the highest quality products. Invivoscribe® warrants that the products meet or exceed the performance standards described in the Instructions For Use, as to products with such an insert. If a product is covered by product specifications and does not perform as specified, our policy is to replace the product or credit the full purchase price. No other warranties of any kind, expressed or implied, are provided by Invivoscribe®. Invivoscribe® liability shall not exceed the purchase price of the product. Invivoscribe® shall have no liability for direct, indirect, consequential or incidental damages arising from the use, results of use, or inability to use its products; product efficacy under purchaser controlled conditions in purchaser’s laboratory must be established and continually monitored through purchaser defined and controlled processes including but not limited to testing of positive, negative, and blank controls every time a sample is tested. Ordering, acceptance, and use of product constitutes purchaser acceptance of sole responsibility for assuring product efficacy and purchaser agreement to the limitation of liability set forth in this paragraph.
This product is for Research Use Only; not for use in diagnostic procedures.
This product is covered by one or more of the following: European Patent Number 1549764, European Patent Number 2418287, European Patent Number 2460889, Japanese Patent Number 4708029, United States Patent 8859748, United States Patent 10280462, and related pending and future applications. All of these patents and applications are licensed exclusively to Invivoscribe®. Additional patents licensed to Invivoscribe® covering some of these products apply elsewhere. Many of these products require nucleic acid amplification methods such as Polymerase Chain Reaction (PCR). No license under these patents to use amplification processes or enzymes is conveyed expressly or by implication to the purchaser by the purchase of this product.
©2024 Invivoscribe, Inc. All rights reserved. The trademarks mentioned herein are the property of Invivoscribe, Inc. and/or its affiliates, or (as to the trademarks of others used herein) their respective owners.

 Mexico and Canada
Mexico and Canada  Outside North America
Outside North America  US
US